Holocentric chromosomes evolution
Chromosomal evolution and its relationship with diversification pattern in angiosperms: a macro and microevolutionary approach through the hyperdiverse family of the sedges
PhD associated with the project funded by the Ministry of Science, Innovation and Universities (PIs: Modesto Luceño and Santiago Martín-Bravo)
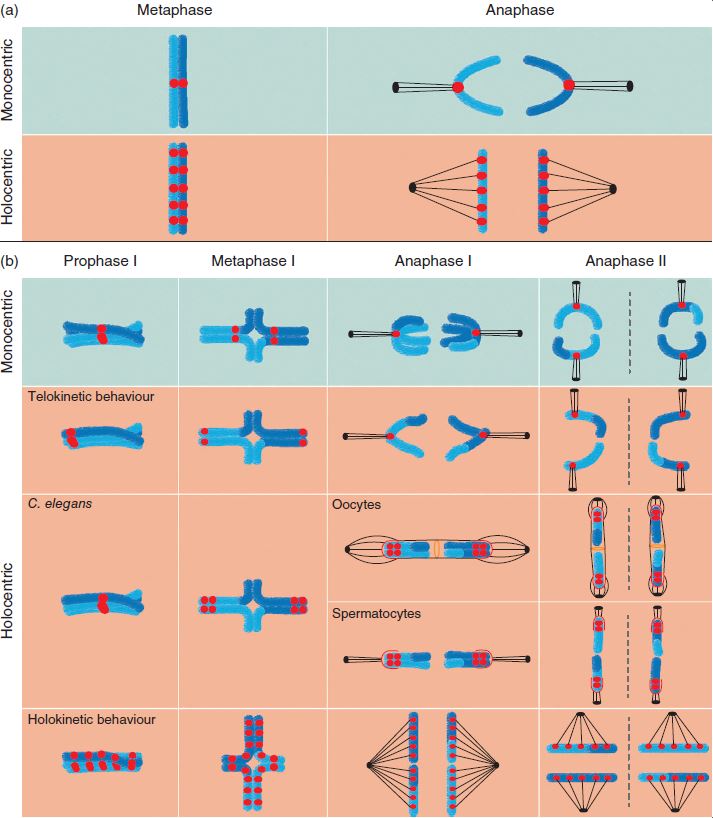
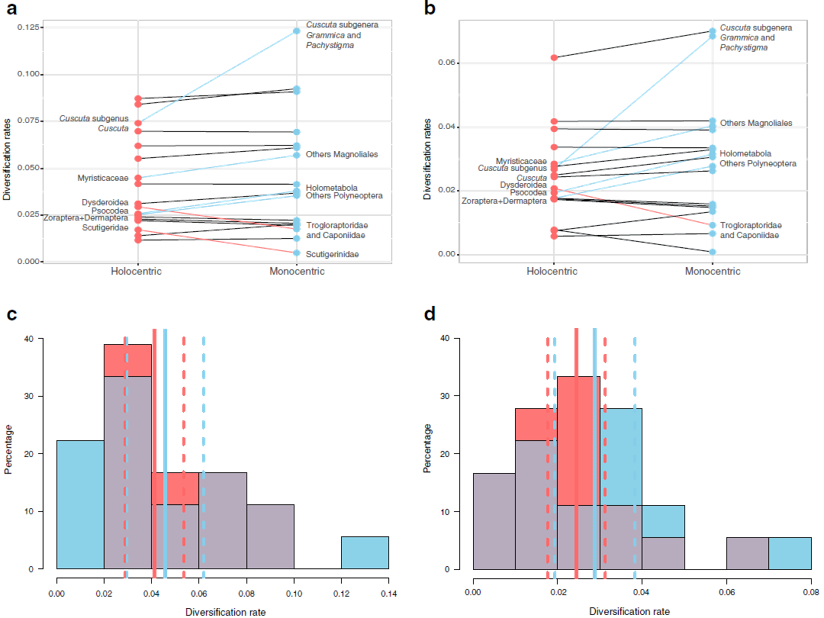
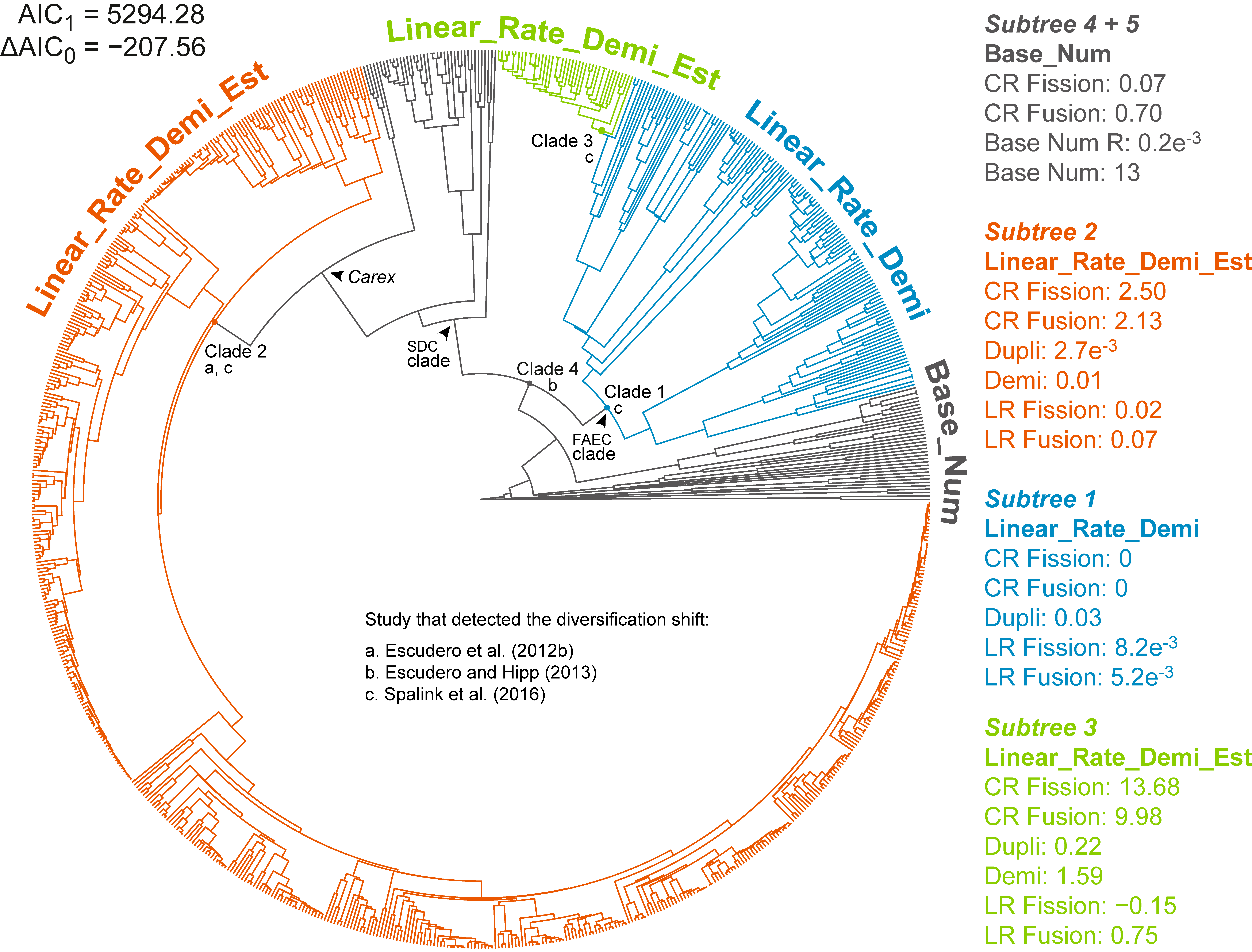
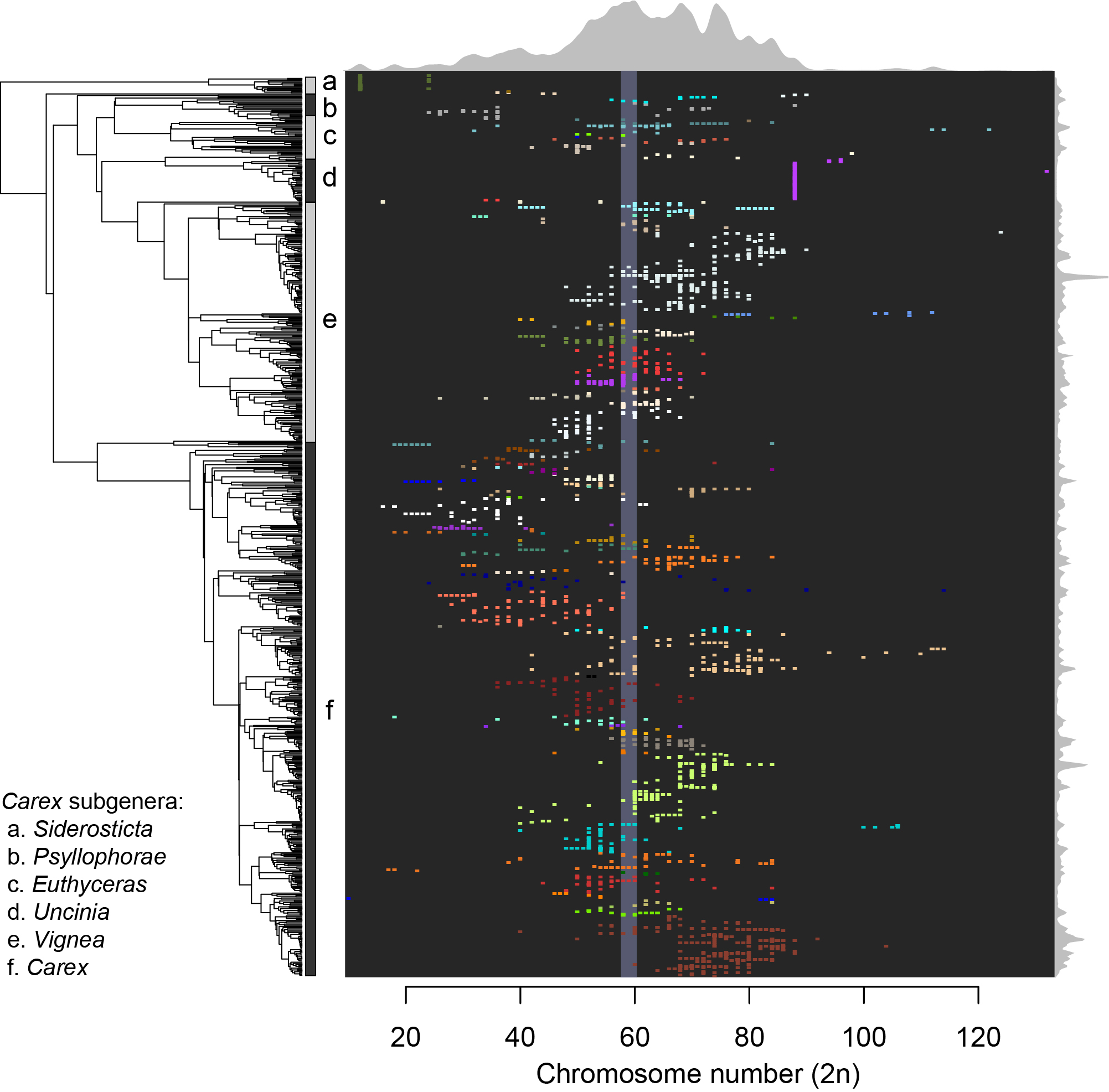
Chromosome evolution is one of the main drivers of lineage differentiation and, eventually, speciation among eukaryotic organisms. This diversification could be triggered either by mutations involving chromosome number change (whole genome duplication –polyploidy– and dysploidy events –fusion or fission–) or by translocations in certain chromosomes. The centromere plays a fundamental role in the cell division process with respect to genetic inheritance. Chromosomes with a single centromere are named monocentric, whilst the socalled holocentric chromosomes present a large number of centromeres throughout the chromosomes. If a holocentric chromosome was to break, remaining fragments would be viable in the subsequent cell division events, due to the fact that each fragment would be carrying at least one functional centromere. In this Ph.D dissertation, we have analysed the relation between holocentric chromosomes and lineage diversification at different evolutionary scales, from domain to genus. In the first place, we have studied the origin of monocentric and holocentric chromosomes in the eukaryotic tree of life , and the transitions between both types, with reversals from holocentry to monocentry retrieved as a more frequent transition than the opposite. In the second place, we have comparatively studied the diversification of holocentric lineages with respect to their monocentric sister lineages, observing that there are no significant differences in the net diversification rates between both types. Next, we descended in the evolutionary scale, to study whether different modes of chromosome number evolution exist in the most speciesrich holocentric plant lineage (family Cyperaceae, order Poales). We found significant support for a complex scenario involving different modes of chromosome evolution in different clades, related with previously reported diversification rate shifts in the family. Finally, we focused on the relationship between chromosome number and several climatic factors and morphological traits in the megadiverse genus Carex (Cyperaceae). This is not only the most diverse among the sedges genera, with ca. 2000 species, but also one of the three most species-rich angiosperm genera. Carex displays an outstanding chromosome number variation (2n = 10–132), and its karyotype evolves mainly by means of dysploidy. We found significantly higher diversification rates around intermedial chromosome numbers within the genus (2n = ca. 58–60). We infer that, despite showing a multitude of distinct evolutionary histories, chromosome number seems to have had an impact on the genus evolution, in part related to morphological and niche climatic characteristics.

 Bionomia
Bionomia